|
|
|
Indian Pediatr 2016;53: 211-215 |
 |
ADRB2 Polymorphism and Salbutamol
Responsiveness in Northern Indian Children with Mild to Moderate
Exacerbation of Asthma
|
|
Puneet Kaur Sahi, Shivaram Shastri, Rakesh Lodha,
Neerja Gupta, #RM Pandey,
Sushil Kumar Kabra and Madhulika Kabra
From Departments of Pediatrics and #Biostatistics,
All India Institute of Medical Sciences, New Delhi, India.
Correspondence to: Dr Madhulika Kabra, Professor,
Division of Genetics, Department of Pediatrics, All India Institute of
Medical Sciences, New Delhi, India.
Email:
[email protected]
|
Objectives: The primary objective was to determine the association
between beta-2 adrenergic receptor (ADRB2) gene
polymorphism (rs1042713, c.46A>G, p.Arg16Gly) and the response to
inhaled salbutamol in North Indian children aged 5 to 15 years, with
mild to moderate exacerbation of asthma.
Methods: This cross-sectional study was done at a
tertiary-care hospital in Northern India from June 2011 to May 2013. 120
children with asthma with mild to moderate exacerbation underwent
spirometry at baseline and after administration of three doses of
salbutamol. An increase in FEV1 ³15%
was considered as positive response. Blood samples from these children
were analysed for ADRB2 polymorphism (p.Arg16Gly). 94
non-asthmatic adult controls were also studied to determine the
prevalence of ADRB2 polymorphism.
Results: In asthmatic children, the frequency of
AA, GG, AG genotypes were 24.2%, 24.2% and 51.7% compared to 20.2%, 20.2
% and 59.6%, respectively in the non-asthmatic adults. Salbutamol
responsiveness showed no correlation with the studied ADRB2
polymorphism (p= 0.55). A trend towards greater bronchodilator
responsiveness amongst AA genotype, compared to GG genotype was observed
(Median change in percent predicted FEV1 14.5% and 7.5%, respectively).
Conclusions: No correlation was found between
salbutamol responsiveness and ADRB2 genotype in Northern Indian
children with asthma with mild-to moderate exacerbation.
Keywords: Adrenergic receptor, Bronchodilation, Management,
Prediction.
|
|
A
sthma affects around 6-31% of Indian children
[1]. Short-acting beta-2 agonists (SABA) form the mainstay of treatment
of acute exacerbations. However, a significant heterogeneity exists in
response to inhaled SABA, of which 70-80% may have a genetic basis [2].
SABA act by binding to the beta 2 adrenergic receptor (ADRB2), coded by
an intron-less gene on chromosome 5. A single nucleotide polymorphism
(SNP) at nucleotide position 46 of this gene (AGA to GGA) substitutes
the 16th amino acid of the
translated protein chain from arginine to glycine and alters receptor
function. This p.Arg16Gly polymorphism has been the subject of intense
research. Conflicting data exist regarding effect of this polymorphism
on salbutamol response particularly with regard to different
ethnicities. There is a paucity of Indian data, especially in children.
Thus, our first objective was to determine an association between
Salbutamol response and p.Arg16Gly polymorphism of ADRB2 gene in
children with asthma with acute exacerbation. The second was to
determine prevalence of these genotypes in the asthmatic and
non-asthmatic population.
Methods
This cross-sectional study was carried out between
June 2011 to May 2013 at a tertiary care hospital in India. The protocol
was approved by the Institute’s ethics committee and written informed
consent was taken from the parents of all participating children, and
from all adults enrolled as controls. Adults were chosen as controls,
considering that children appearing healthy currently might develop
asthma symptoms over a period of time.
Calculation of sample size was based on the
observation that compared to homozygotes for Gly-16, homozygotes for
Arg-16 were 5.3 times more likely to respond to salbutamol [3]. The
prevalence of the Arg16 polymorphism in the general population is shown
to be 30-50% [4]. With 50% precision, confidence interval of 95%, sample
size was calculated to be 120 [5]. For controls, we planned to enroll
120 adults.
Children between 5-15 years with physician- diagnosed
asthma (three or more episodes of reversible airway obstruction
documented by bronchodilator response) were screened for enrolment.
Those with mild to moderate acute exacerbation, as determined by the
Clinical Asthma Score (CAS) [6] were included. Children with life
threatening asthma, pre-existing chronic medical conditions, use of
long-acting beta agonists in prior 2 weeks, salbutamol therapy by any
route in 6 hours preceding the evaluation, and those on oral steroid
therapy were excluded.
Clinical details and CAS were recorded in a
pre-designed structured proforma. All enrolled children underwent
spirometry using a portable spirometer (Superspiro MK2 Micro Medical
Ltd, UK), as per standard technique. The absolute and percentage
predicted values of following parameters were recorded: FEV 1 (Forced
Expiratory Volume-1 second); FVC (Forced Vital Capacity) PEFR (Peak
Expiratory Flow Rate) FEF50 (Forced
Expiratory Flow 50). Thereafter, they received salbutamol 100 µg 2 puffs
with MDI (metered dose inhaler) with spacer every 10 min for a total of
three doses followed immediately by repeat spirometry. The highest FEV
which was available from all adequate curves produced during baseline
and post treatment spirometry were recorded. Percentage increase in
actual FEV was measured using the formula [(FEV (post-bronchodilator) –
FEV baseline)/FEV baseline] X 100. Positive bronchodilator response was
considered if percentage increase in FEV1
was ³15%.
For DNA analysis, 3-5 mL blood was collected in EDTA
vacutainers and stored at 4ºC. DNA was extracted using the phenol
chloroform method [7]. A 168 bp region flanking the p.Arg16Gly
polymorphism region was amplified using Polymerase Chain reaction (PCR).
The forward primer: 5' GCC TTC TTG CTG GCA CCC CAT 3' (21 bases)
and reverse primer: 5' CAG ACG CTC GAA CTT GGC CAT G 3' (22
bases) were used. PCR reactions were carried out in 25 µL mix containing
2.5 µL of 10X PCR buffer, 2.5 µL of 2 mM dNTPs, 1.0 µL of 10 µM forward
and reverse primer each, 0.25 µL of 3 U/µL Taq polymerase, 16.75
µL of sterile water and 1 µL of extracted DNA (100 ng/mL). The above mix
was kept in a thermal cycler with set temperature conditions repeated
for 35 cycles. Initial denaturation was done at 94 ºC
for 2 minutes followed by denaturation at 94ºC
for 40 seconds, annealing at 64ºC for 40 seconds, extension at 72ºC
for 50 seconds and final extension at 72ºC for 5 minutes. A blank, with
all components except the DNA template, was run simultaneously with each
run of PCR as control. The PCR products were checked for adequate DNA
amplification by a run at 150 volts in a horizontal gel electrophoresis
system. The underlined bases in both the forward and reverse primers
were modified from A to C to create restriction sites for the NcoI
enzyme which was later used in restriction digestion of the PCR
products. The forward primer creates an NcoI restriction site on
Gly16 PCR product but not on Arg16 PCR product. The reverse primer
creates a restriction site on both, thus serving as a control to assess
whether restriction digestion was complete.
The amplified PCR products were subjected to
restriction digestion with NcoI enzyme. 8.8 µL of PCR amplified
DNA, 1.0 µl of 1X NE buffer 3, 0.25 µL of NcoI enzyme (10,000 U/mL)
were mixed and incubated at 37ºC for 16 hours. The products of
restriction digestion along with a control (unrestricted PCR product)
were run at 150 volts in a horizontal electrophoresis system on a 4%
agarose gel. DNA ladder was run to judge the size of the cut products.
The gel was observed under UV light and the image was stored.
After restriction digestion of the PCR products of
168 bp: Homozygote AA give 146 bp and 22 bp products; Heterozygotes AG
give 146 bp, 128 bp, 22 bp and 18 bp products; Homozygotes GG give 128
bp, 22 bp and 18 bp products.
Data were collected in a structured proforma and
managed using MS Excel software. Statistical analysis was done using
STATA 11. The comparison of salbutamol responsiveness of children with
different ADRB2 genotypes (AA/AG/GG) was done using the Pearson
Chi square test. P value
less than 0.05 was considered statistically significant. Hardy-Weinberg
equilibrium analysis of ADRB2 SNP at 46th
nucleotide position was also performed [8].
Results
During the study period, 246 children were screened
for inclusion in to the study. Of these, 120 results were finally
available for analysis (Fig. 1). The baseline
characteristics of the study population are tabulated in Table
I.
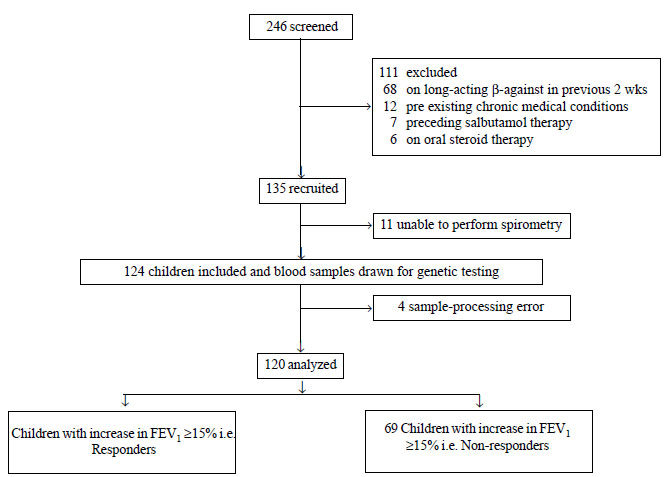 |
|
Fig. 1 Study flow for screening,
enrolment and stratification into responders and non-responders.
|
Of the analyzed 120 children with asthma, 42.5% were
labelled as responders. Comparison of responders and non-responders
shows that they were are ell-matched in characteristics which could have
confounded the response to salbutamol (Table I).
TABLE I Baseline Charactersitics of Responders Versus Non Responders
|
Variable |
Responders |
Non- |
P |
|
|
(n=51) |
responders |
value |
|
|
(n=69) |
|
|
Age; mean (SD), y |
9.3 (2.12) |
9.1 (2.37) |
0.76 |
|
Female gender, n (%) |
11 (21.6) |
18 (26.1) |
0.56 |
|
Height; mean (SD), cm |
131.3 (13.1) |
131.0 (15.3) |
0.90 |
|
Weight; mean (SD), kg |
27.08 (8.47) |
26.89 (10.1) |
0.91 |
|
Age at onset; mean (SD), mo |
46.11 (36.6) |
55.95 (42.4) |
0.19 |
|
Severity asthma, n (%) |
|
|
0.85 |
|
Intermittent |
06 (11.7 ) |
09 (13.0) |
|
|
Mild persistent |
26 (50.9 ) |
30 (43.5) |
|
|
Moderate persistent |
18 (35.3 ) |
29 (42.0) |
|
|
Severe persistent |
01 (01.9 ) |
01 (01.4) |
|
|
Family history of allergy, n (%) |
42 (60.9 ) |
35 (68.6) |
0.38 |
No significant association was found between the
ADRB2 genotypes and salbutamol-responsiveness (P= 0.55) (Table
II). However, a trend towards greater bronchodilator
responsiveness was seen amongst those who were carrying the A/A
polymorphism as compared to G/G homozygotes. The median of change in
percentage predicted FEV1 in the three ADRB2 genotypes was 14.5
for A/A, 12.5 for A/G and 7.7 for G/G genotypes, respectively.
TABLE II ADRB2 Genotype and Bronchodilator Response in the Study Population
|
ADRB2 Genotype |
Responders |
Non-responders, |
|
no. (%), n = 51 |
no. (%), n = 69 |
|
A/A |
14 (27.4 ) |
15 (21.7 ) |
|
A/G |
27 (52.9 ) |
35 (50.7 ) |
|
G/G |
10 (19.6 ) |
19 (27.5 ) |
|
P values for comparisons between A/A vs G/G, A/G vsG/G, A/A
vs A/G and G/G vs A/A + A/G were all >0.05. |
No significant association was found between the
studied ADRB2 genotypes and the severity of asthma (P= 0.39) or
family history of asthma (P=0.25). In the 120 children with
asthma, the frequency of AA, GG, AG genotypes had prevalence of 24.2%,
24.2% and 51.7% respectively and were in equilibrium as per the Hardy
Weinberg law ( c2=
0.13).
One hundred fourteen non-asthmatic adults were
enrolled as controls to determine prevalence of ADRB2 genotypes.
Twenty samples were lost to processing errors, leaving 94 for analysis.
Mean age of control population was 42.5 yrs (39.3% females). The
prevalence of ADRB2 genotypes was 20.2%, 59.6%, and 20.2% for
A/A, A/G and G/G, respectively which fall within the Hardy Weinberg
equilibrium.
No ADRB2 genotype was found more prevalent in
the asthmatic population compared to the non-asthmatics (P=0.51).
The allele frequencies were found to be exactly equal at 50.0% for wild
type A allele as well as the mutant G allele in both the asthmatic and
non-asthmatic groups.
Discussion
In this observational study done amongst children
with asthma aged 5 to 15 years with acute exacerbation of asthma, no
association was found between the ADRB2 genotype (Arg16Gly) and
salbutamol-responsiveness defined as the percentage change in FEV 1
³15%.
Green, et al. [9] suggested that several SNPs
of the ADRB2 gene
significantly alter ADRB2 receptor down regulation.
Unfortunately, the multitude of studies subsequently done have shown
vastly discordant results. Early studies by Martinez, et al. [3]
showed better bronchodilator response in asthmatics with A/A genotype of
ADRB2 compared to the G/G genotype (P=0.05). Later,
similar results were seen in several studies [10-12]. A study by
Choudhary, et al. [13] showed better salbuta-mol response in
those with AA genotype in Puerto Ricans but not in Mexican highlighting
ethnic differences. In contrast a few studies have shown absolutely
opposite results [14]. The only Indian study done showed a better
salbutamol response in those possessing the G/G ADRB2 genotype
[15]. Some larger studies have reported lack of any such association
[16-20].
This study is one of the few studies which have
researched the response to inhaled SABA during an acute exacerbation of
asthma. Most other studies have focused on cohorts of stable asthma
patients [3,10-12,15,16,18]. This has direct implications for finding
the best personalized treatment for acute asthma attacks that are
responsible for hospitalization and mortality. Percentage change in
actual FEV was chosen as the main outcome measure as it is the most
objective and immediate outcome. It has been taken as the study
end-point in several studies, making the comparison of results easier. A
suitably high cut-off of ³15%
was taken as a meaningful response to rule out any measurement
variability [21]. Though no significant association could be established
in the current study, a trend towards a greater positive bronchodilator
response amongst the Arg16Arg homozygotes has been seen. A larger sample
size could prove or disprove such an association. Importantly, the
results of our study have not shown concordance with the only other
Indian study on the subject [15], though it involved a Southern Indian
population. This ethnic difference of the populations enrolled may
explain the variability in the results obtained, highlighting the need
of conducting these studies in various ethnic groups.
There are certain inherent limitations to our
interpretations. The foremost is the fact that there are a multitude of
SNPs of ADRB2 gene and many of them may be in linkage
disequilibrium, meaning that a certain set of alleles are more likely to
be inherited together as a block. Thus the protective effect of one SNP
may mask the adverse effect of another SNP when inherited together.
Hence, research studying association of ADRB2 haplotypes with
bronchodilator response may be more relevant. The other concern is that
extrapolation of these results to the Indian population warrants a
larger sample size, with due importance given to the various
ethnicities. Even though we have restricted our study to the northern
Indian population, the region within itself contains diverse population
groups requiring studies focussed on these subgroups. Thirdly, the study
involved only two cases of severe persistent asthma which may possess
altogether different genotypic and clinical manifestations compared to
the less severe variants of asthma. Hence, more studies need to be
conducted before drawing conclusions specifically in this group.
Contributors: PS: planning of study, data
collection and analysis, laboratory work and writing of manuscript, SS:
involved in planning of study, laboratory work and manuscript writing,
RL: planning of study, data collection and analysis, and writing of
manuscript, NG: data collection, writing of manuscript; RMP and SKK:
planning of study, data analysis and writing of manuscript, MK: study
planning, data collection, laboratory analysis, and manuscript writing.
MK: will act as guarantor for the study.
Funding: None; Competing interests: None
stated.
|
What is Already Known?
•
Polymorphism of ADRB2 gene on chromosome 5 may
influence the salbutamol response in acute asthma exacerbation.
What This Study Adds?
•
Salbutamol responsiveness showed no correlation with the
studied ADRB2 genotypes.
|
References
1. Paramesh H. Epidemiology of asthma in India.
Indian J Pediatr. 2002;69:309-12.
2. Drazen JM, Silverman EK, Lee TH. Heterogeneity of
therapeutic responses in asthma. Br Med Bull. 2000;56:1054-70.
3. Martinez FD, Graves PE, Baldini M, Solomon S,
Erickson R. Association between polymorphisms of ADRB2 and response to
albuterol in children with and without history of wheezing. J Clin
Invest. 1997;100:3184-88.
4. Hall IP, Blakey JD, Al Balushi KA, Wheatley A,
Sayers I, Pembrey ME, et al. Beta 2-adrenoceptor polymorphisms
and asthma from childhood to middle age in the British 1958 birth
cohort: a genetic association study. Lancet. 2006;368:771-9.
5. Lemeshow S, Hosmer DW Jr, Klar J, Lwanga SK.
Tables for sample size determination, table 9h In: Lemeshow S,
Hosmer DW Jr, Klar J, Lwanga SK. Adequacy of Sample Size in Health
Studies. England, John Wiley & Sons Ltd, 1990. p. 156.
6. Carroll CL, Sekaran AK, Lere RJ, Schramm CM. A
modified Pulmonary index score with predictive value for pediatric
asthma exacerbations. Ann Allergy Asthma Immunol. 2005;94;355-9.
7. Sambrook J, Russell DW. Isolation of HMW DNA from
mammalian cells using proteinase K and phenol, protocol 1. In: Sambrook
J, Russell DW. Molecular Cloning: A Laboratory Manual. Third edition;
volume 1. New York, Cold Spring Harbor laboratory Press, 2001. p. 6.8.
8. Hartl DL, Clarke AG. Principles of Population
Genetics. Sunderland, MA: Sinauer, 2007. p. 48-54.
9. Green SA, Cole G, Jacinto M, Innis M, Liggett SB.
Polymorphism of human ADRB2 alters functional properties of
receptor. J Biol Chem. 1993;268:23116-21.
10. Kotani Y, Nishimura Y, Maeda H, Yokoyama M.
Beta2-adrenergic receptor polymorphisms affect airway responsiveness to
salbutamol in asthmatics. J Asthma. 1999;36:583-90.
11. Cho SH, Oh SY, Bahn JW, Choi JY, Chang YS, Kim
YK, et al. Association between bronchodilating response to SABA
and non-synonymous SNP of ADRB2 gene. Clin Exp Allergy.
2005;35:1162-7.
12. Salah K, Morsy S, Atta A. Effects of ADRB2
polymorphism on asthma severity and response to salbutamol on Egyptian
children. Egypt J Pediatr Allergy Immunol. 2012;10:81-6.
13. Choudhry S, Ung N, Avila PC, Ziv E, Nazario S,
Casal J. Pharmacogenetic differences in response to albuterol between
Puerto Ricans and Mexicans with asthma. Am J Respir Crit Care Med.
2005;171:563-70.
14. Carroll CL, Stoltz P, Schramm CM, Zucker AR.
ADRB2 polymorphisms affect response to treatment in children with
severe asthma exacerbations. Chest. 2009;135:1186-92.
15. Kukreti R, Bhatnagar P, B-Rao C, Gupta S, Madan
B, Das C, et al. Beta(2)-adrenergic receptor polymorphisms and
response to salbutamol among Indian asthmatics. Pharmacogenomics.
2005;6:399-410.
16. Tsai HJ, Shaikh N, Kho JY, Battle N, Naqvi M,
Navarro D, et al. ADRB2 polymorphisms: Pharmacogenetic
response to bronchodilator among African American asthmatics. Hum Genet.
2006;119:547–57.
17. Martin AC, Zhang G, Rueter K, Khoo SK, Bizzintino
J, Hayden CM, et al. ADRB2 polymorphisms predict response
to beta2-agonists in children with acute asthma. J Asthma.
2008;45:383-8.
18. Szczepankiewicz A, Breborowicz A, Sobkowiak
P, Kramer L, Popiel A. Role of ADRB2
gene polymorphism in asthma and response to beta2-agonists in
Polish children. J Appl Genet. 2009;50:275-81.
19. Fu WP, Zhao ZH, Zhong L, Sun C, Fang LZ, Liu L,
et al. Relationship between polymorphisms in the 5 leader cistron,
positions 16 & 27 of ADRB2 gene & asthma in a Han population from
southwest China. Respirology. 2011;16:1221-7.
20. Isaza C, Sepúlveda-Arias JC, Agudelo BI,
Arciniegas W, Henao J, Porras GL, et al. ADRB2 polymorphisms in
asthmatic and non-asthmatic school children from Colombia and their
relationship to treatment response. Pediatric Pulmonol. 2012; 47:848-55.
21. Nickerson BG, Lemen RJ, Gerdes CB, Wegmann MJ,
Robertson G. Within-subject variability and per cent change for
significance of spirometry in normal subjects and in patients with
cystic fibrosis. Am Rev Respir Dis. 1980;122:859-66.
|
|
|
 |
|

