From the Divisions of Genetics and Neonatology,
Department of Pediatrics, All India Institute of Medical Sciences, New
Delhi 110 029, India.
Correspondence to: Dr. Madhulika Kabra, Associate
Professor, Department of Pediatrics, All India Institute of Medical
Sciences, New Delhi 110 029.
E-mail: madhulikakabra@hotmail.com
Manuscript received: July 8, 2002; Initial review
completed: August 13, 2002; Revision
accepted: December 23, 2002
Spinal Muscular atrophy (SMA) Type I is a fatal
autosomal recessive disease caused by homozygous deletion of telometric
region of exon 7/8 of the SMN gene. Prenatal diagnosis is feasible and
desirable by most families. We report on prenatal diagnosis of SMAI in a
family where dried umbilical cord stump from the deceased affected baby
was used to confirm the diagnosis. Prenatal diagnosis was provided in
the subsequent pregnancy. We emphasize the need for storing DNA from
individuals affected with suspected single gene disorders.
Key words:
Prenatal diagnosis, spinal muscular atrophy Type I.
Extraction of DNA from old forensic specimens for
genetic analysis is a known technique and has found wide application in
forensic medicine and pathological investiga-tions(1,2). We would like
to highlight an interesting case where it was possible to retrieve DNA
from umbilical cord of a baby who died of suspected spinal muscular
atrophy (SMA) 2 years ago. Retrieval of DNA was helpful in prenatal
diagnosis of SMA in subsequent pregnancy.
Case Report
A 33-year-old 4th gravida mother, resident of Uttar
Pradesh, with no previous live issue, was referred to us early in the
2nd trimester for antenatal dianosis of a possible neuromuscular
disorder. Her first pregnancy had resulted in a spontaneous abortion.
Both her subsequent pregnancies had resulted in the birth of floppy
infants with neuromuscular weakness. The babies had difficulty in
feeding and breathing, and died in infancy. Both the infants were
treated at a local hospital and no definite diagnosis was avail-able as
no neurophysiological/pathological studies were done, however clinical
picture described by parents was suggestive of a neuromuscular disorder
with a strong possibility of SMA type I. Parents were keen to have a
healthy baby which was possible only by prenatal testing. Since genetic
testing was not performed on the previous babies, no DNA from affected
children was available for the exclusion of SMA in this pregnancy.
However, due to an age-old custom, the umbilical cord of the last
sibling was still available. In certain parts of rural India, the fallen
off umbilical stump is tied by the bedside for the first 6 weeks and
later sacredly buried among the roots of a Banyan/Bamboo tree. In their
anxiety for the last sibling, the parents had forgotten to bury the cord
and it still hung by the bedside even after two years. We thought of
trying to retrieve DNA from the old cord. The parents were able to get
the dried remains of the cord from which DNA was extracted successfully.
The cord was cut in thin pieces and washed with Tris EDTA (TE) in 1.5 mL
eppendrof tube. 500 µL of TE, 40 µL of 20% SDS and 10 µL of proteinase K
(200 µL/mL) was added and kept at 65ºC for 2 hrs. After this 10µL of
proteinase K was added again, mixture was vortexed and kept at 37ºC for
48 hrs. The mixture was then microcentrifuged for 5 minutes at 5000 rpm
at room temperature. DNA extraction was done using the standard phenol
chloroform extraction method twice(3). 3 M sodium acetate and ethanol
were added mixed by inversion twice and microcentrifuged for 5 minutes
and supernatant was recouped. The pallet was washed with 70% ethanol,
dried and resuspended in distilled water and used for polymerase chain
reaction (PCR).
The PCR method and conditions used for amplifying
exon 7 and 8 of SMN genes used were as described by Shuan-Peilin(4). The
PCR products were digested by restriction enzyme Dra 1 and Dde 1 for
exon 7 and exon 8 respectively and then electrophoresed on 3.5% agarose
gel.
On evaluation parents showed two fragments of exon 7
after Dra 1 digestion whereas in umbilical cord DNA (i.e.,
affected child’s) telomeric copy was deleted (Fig.
1). This confirmed the diagnosis of SMA in the previous
child. CVS DNA had an intact telomeric copy whereas centromeric copy was
deleted. Similarly, exon 8 telomeric deletion was found in the cord DNA
whereas CVS DNA did not show telomeric deletion (Fig.1). Parents
were counseled and they decided to continue with the pregnancy. A
healthy term female baby was born. She is in regular follow up and is
asymptomatic at 6 months of age. The test was repeated postnatally and
it matched with the CVS results.
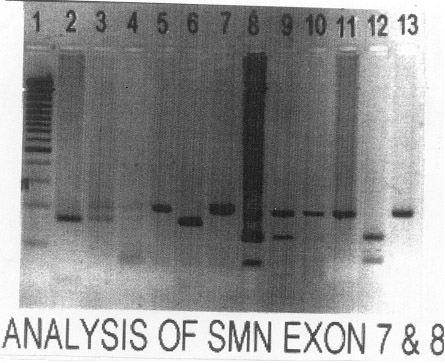 |
|
Fig. 1. Analysis of SMN exon 07 (lane 2-7) and
8 (lane 8-13). Lane 1: 100 bp ladder; Lane 2: +ve control for exon
7 telomeric deletion; Lane 3: mother; Lane 4: father; Lane 5: CVS
(+ve for centromeric deletion); Lane 6: umbilical cord DNA (+ve
for telomeric deletion); Lane 7: baby at the age of 3 months (+ve
for centromeric deletion); Lane 8: –ve control for exon t; Lane 9:
mother; Lane 10: father; Lane 11: CVS (+ve for centromeric
deletion); Lane 12: umbilical cord DNA (+ve for telomeric
deletion); Lane 13: baby at the age of 3 months (+ve for
centromeric deletion).
|
Discussion
The gene for SMA (all three types) has been mapped to
chromosome 5q13(5). Out of two candidate genes, i.e., survival
motor neuron gene (SMN) and neuronal apoptosis inhibitory gene (NAIP),
SMN gene has been implicated more commonly as the causative gene.
According to Western literature in about 95-98% of SMA I patients there
is homozygous deletion of the telomeric copy exon 7 of SMN gene(6).
Isolated homo-zygous deletion of the centromeric copy does not lead to
symptoms, though recently a case report from India has described one
such SMA case(7). Homozygous centrometric deletion has been reported in
normal asymptomatic individuals also(6). Presence of centromeric
deletion in addition to telo-meric deletion/point mutation can probably
contribute to severity of disease. In the present family parents were
counseled about all these aspects and they decided to continue with the
pegnancy.
Literature review has shown that DNA extraction is
possible even years later from postmortem tissue. DNA has been extracted
and purified from various forensic specimens(2,8). DNA material has been
extracted successfully from specimens even after prolonged periods
ranging from a few weeks to many months. Some studies have shown that
there is no correlation between the age of the specimen and the extent
of DNA preservation(2). Specific gene fragments can be amplified for
sequencing or fingerprinting. This has found wide applications in the
field of forensic medicine but rearely in clinical set-up. In our case
it was possible to extract DNA from an old umblical cord. Genetic
studies for SMA from this DNA were helpful in the antenatal diagnosis
and genetic counseling.
As the DNA testing of affected individual is crucial
for diagnosis and prenatal diagnosis of single gene disorders, it is
advisable that the physicians taking care of children with suspected
genetic disorder save blood for DNA extraction. Non-availability of the
DNA can be extremely problematic as exemplified by the difficulties
encountered in this case.
Contributors: SA did the molecular studies. MK
was involved in prenatal and postnatal counselling and preparation of
the manuscript and will act as a guarantor of the manuscript. AM and RA
were involved in the evaluation of the baby after birth and preparation
of the manuscript.
Funding: None.
Competing interests: None stated.
